CellChat:细胞相互作用分析
CellChat使用配体受体对应的基因表达对来量化细胞间的相互作用。同时作者也开发了一个CellChat网页版浏览器
具体原理可参见文献,1

CellChat流程
CellChat需要两个输入文件,一个是细胞的基因表达数据;一个是细胞的meta信息。基因表达矩阵要求的是均一化的数据Seurat@assay$RNA@data。如果是counts数据,可以用normalizeData函数来均一化。
首先安装CellChat,以及进行输入数据的准备。从Seurat对象中提取CellChat的输入文件。
rm(list = ls())
options(stringsAsFactors = F)
#devtools::install_github("sqjin/CellChat",force = TRUE)
suppressPackageStartupMessages({
library(Seurat)
library(CellChat)
library(SeuratData)
library(ggplot2)
library(svglite)
library(ggalluvial)
})
try(attachNamespace('pbmc3k.SeuratData'),silent = TRUE)
data("pbmc3k.final")
data.pbmc <- pbmc3k.final@assays$RNA@data
cellmeta <- data.frame(group = pbmc3k.final$seurat_annotations, row.names = names(pbmc3k.final$seurat_annotations)) 创建CellChat对象,进行数据的预处理
# create cellchat abj
cellchat <- createCellChat(object = data.pbmc)
# add cellmeta
cellchat <- addMeta(cellchat,meta = cellmeta,meta.name = "labels")
# set "labels" as default cell identity
cellchat <- setIdent(cellchat,ident.use = "labels")
# table(cellchat@idents)根据物种信息选取不同的数据库,支持human,mouse和zebrafish;CellChat 数据库记录了通路的受体和配体,以及不同类型的细胞相互作用方式,包括:Secreted Signaling,ECM-Receptor,Cell-Cell Contact。
# import cellchat database
CellChatDB <- CellChatDB.human
# Use Cell-Cell Contact for cellchat
CellChatDB.use <- subsetDB(CellChatDB,search = "Cell-Cell Contact",key = "annotation")
# set the used database in the object
cellchat@DB <- CellChatDB.use在这一步,首先提取cellchat中的信号基因,然后鉴定过表达的配体和受体,然后将基因表达数据投射到PPI网络上,为接下来相互作用的推断做铺垫;
# pre processdata
cellchat <- subsetData(cellchat)
cellchat <- identifyOverExpressedGenes(cellchat)
cellchat <- identifyOverExpressedInteractions(cellchat)
cellchat <- projectData(cellchat,PPI.human)推测细胞间在信号通路水平上的通讯,推测的每个配体-受体对的细胞间通信网络和每个信号通路分别存储在“net”和“netP”槽。显著性的配体受体在cellchat@LR$LRsig。
# Interaction Inference
cellchat <- computeCommunProb(cellchat)
cellchat <- computeCommunProbPathway(cellchat)
cellchat <- aggregateNet(cellchat)
# Result
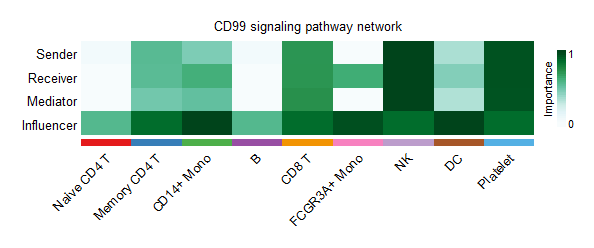
head(cellchat@LR$LRsig)识别细胞群的信号转导作用,通过计算每个细胞群的网络中心性指标,CellChat允许随时识别细胞间通信网络中的主要发送者、接收者、调解者和影响者。
cellchat <- netAnalysis_computeCentrality(cellchat,slot.name = "netP")识别分泌细胞外向(outgoing)交流模式和识别目标细胞的传入(incoming)通信模式。pattern参数一般默认为5.
cellchat <- identifyCommunicationPatterns(cellchat,slot.name = "netP",pattern = "outgoing",k = 5)
outgoing
cellchat <- identifyCommunicationPatterns(cellchat,slot.name = "netP",pattern = "incoming",k = 5)
incoming
结果的可视化
分层图中注意vertex.receiver第一个层次结构图中的目标的细胞组的索引.
# Visualization
# hierarchy
netVisual_aggregate(cellchat, signaling = c("CD99"), vertex.size = 5,layout = "hierarchy",vertex.receiver = seq(1,4),pt.title=20,vertex.label.cex = 0.4)
# circle
netVisual_aggregate(cellchat, signaling = c("CD99"), layout = "circle", vertex.size = 5)
# contribution
#netAnalysis_contribution(cellchat, signaling = c("CD99"))
#netAnalysis_signalingRole_heatmap(cellchat,pattern = "incoming",signaling = "CD99")
netAnalysis_signalingRole_network(cellchat,width = 12, height = 2.5, font.size = 10,signaling = "CD99")
netAnalysis_river(cellchat,slot.name = "netP",pattern = "outgoing")
netAnalysis_river(cellchat,slot.name = "netP",pattern = "incoming")
netAnalysis_dot(cellchat,slot.name = "netP",pattern = "outgoing",dot.size = 5)
netAnalysis_dot(cellchat,slot.name = "netP",pattern = "incoming",dot.size = 5)层次图中,实体圆和空心圆分别表示source和target。圆圈越大细胞数目越多,边缘颜色与信号源一致。线越粗,信号越强。
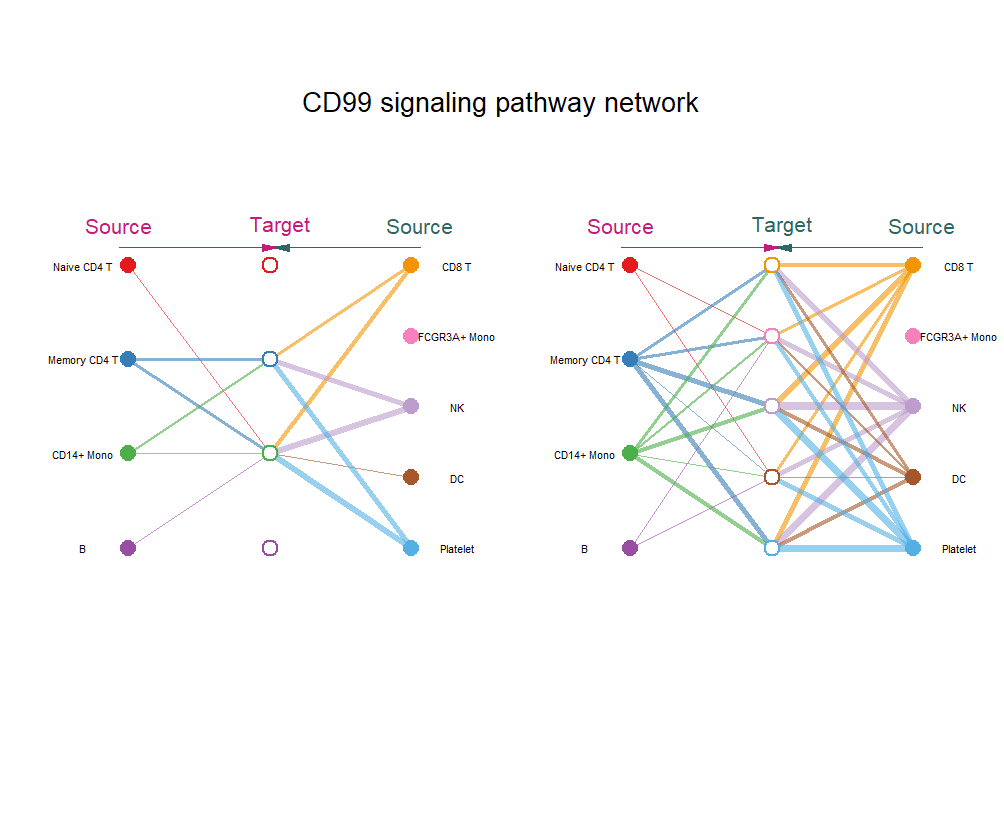
经典的配受体圈图:

识别细胞间通信网络中的主要发送者、接收者、调解者和影响者。
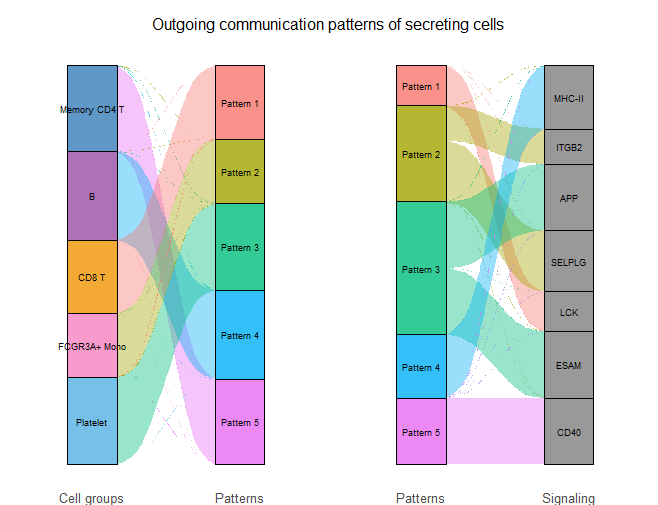
river_outgoing

river_incoming
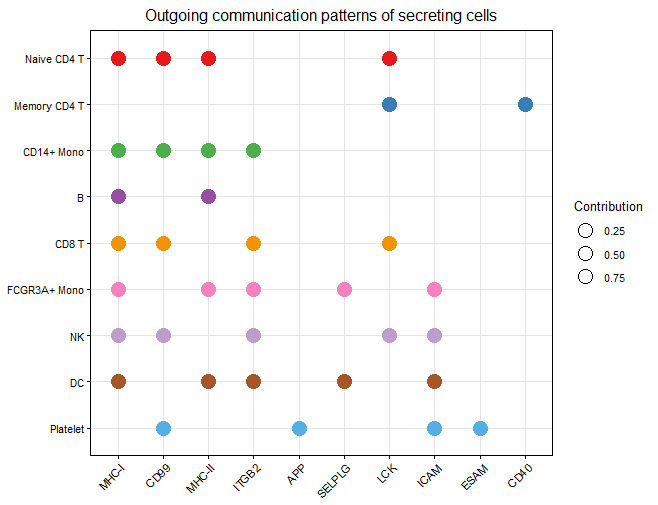
dot_outgoing

dot_incoming
Ref
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nature communications 12, 1088 (2021).↩︎